-
Notifications
You must be signed in to change notification settings - Fork 34
Fr4NK
In a metagenomic sequencing library from a Peruvian vampire bat (Desmodus rotundus, SRA:ERR2756788), we identified a putative Alphoacoronavirus, via a partial sequence match at 93.4% identity to the RdRP of Bat coronavirus Trinidad/1CO7BA/2007 (EU769558).
Assembly of this sequencing library with coronaSPAdes yielded a 29264 nt viral genome. This is a new species of Coronavirus based on RdRP, nucleprotein, membrane protein and replicase 1a, which all classify this virus an Alphacoronavirus outside of all named sub-genera and most similar to a Pedacovirus.
The author of the dataset had previously reported this virus as "DesRot/Peru/AMA_L_F” (Berngner et al 2019), but its sequence is unpublished to the best of our knowledge and was not in our search database.
This is a clear example of a novel CoV within the Serratus data, there are likely scores more of these cases which we are actively working to uncover. Novel members of other virus families can also be identified with this workflow. The summary report for Fr4NK generated by Serratus shows how Fr4NK was identified plus hits to other viruses in the same dataset.
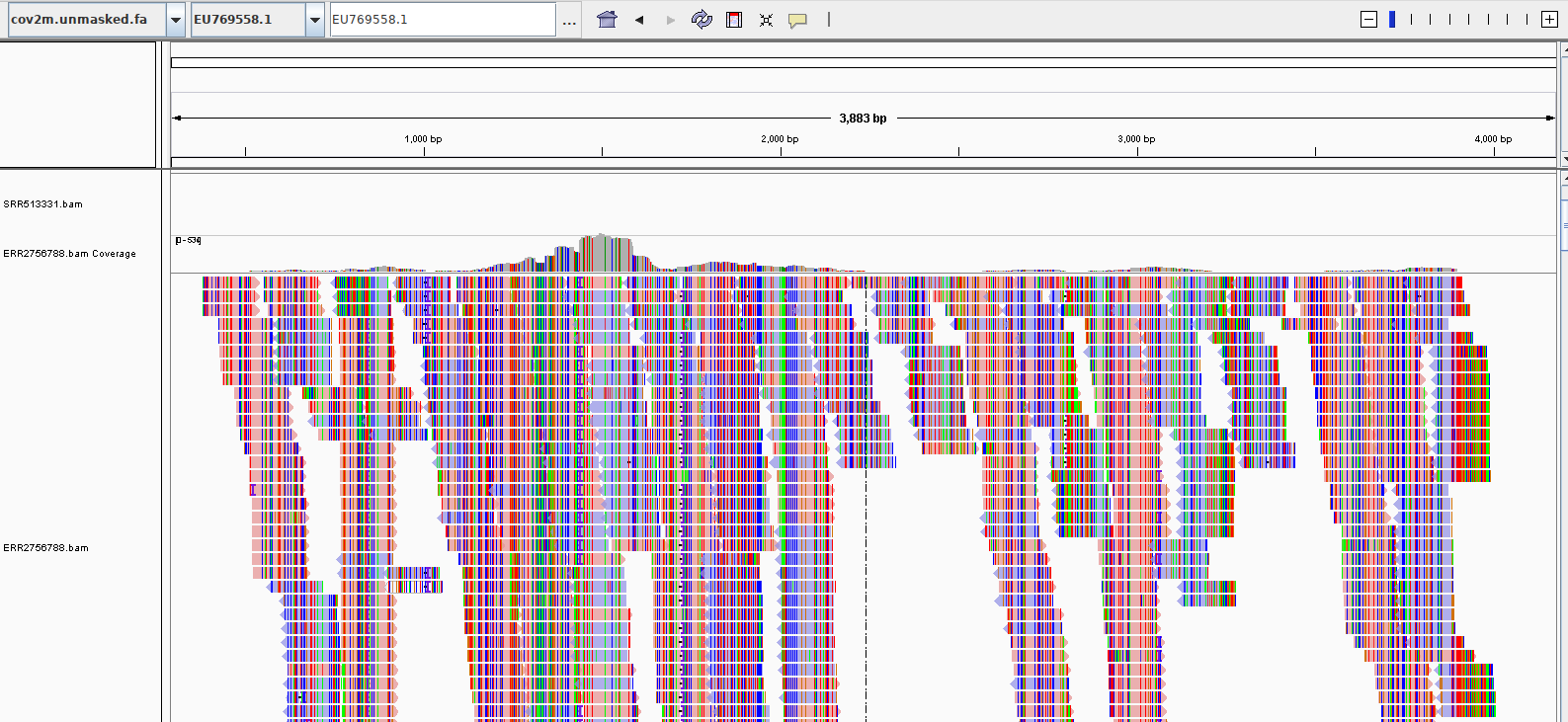
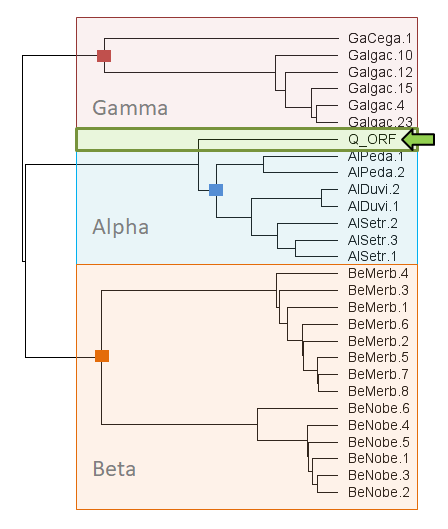
Generated with Serratax, the Serratus taxonomy prediction module, the green arrow indicates the 'query' sequence, which in this case is Fr4NK.
