-
Notifications
You must be signed in to change notification settings - Fork 34
Find_novel_viruses_A_serratus.io
[15 minutes] The easiest way to navigate to a novel virus is to use the serratus.io graphical web interface. This provides a high-level overview of the datasets in the form of aggregated .summary reports.
- Web browser
1. Navigate to serratus.io - RdRP Explorer.
We will perform a Family-level search for Flaviviridae.
There are three controls:
-
Reference Family Selection: This is based on the rdrp1 sequence query. Roughly is is known-virus sequences clustered at ~45% identity and using minimum-linkage.
-
Alignment Identity: The average alignment-identity of all reads in a sequencing dataset mapped to the Reference Family.
-
Score: Is a heuristic score accounting for the number of mapped reads and the span-coverage of a reference sequence. A >80 will often yield a complete RdRP, >50 will often yield a partial RdRP, >10 may yield a partial RdRP sequence.
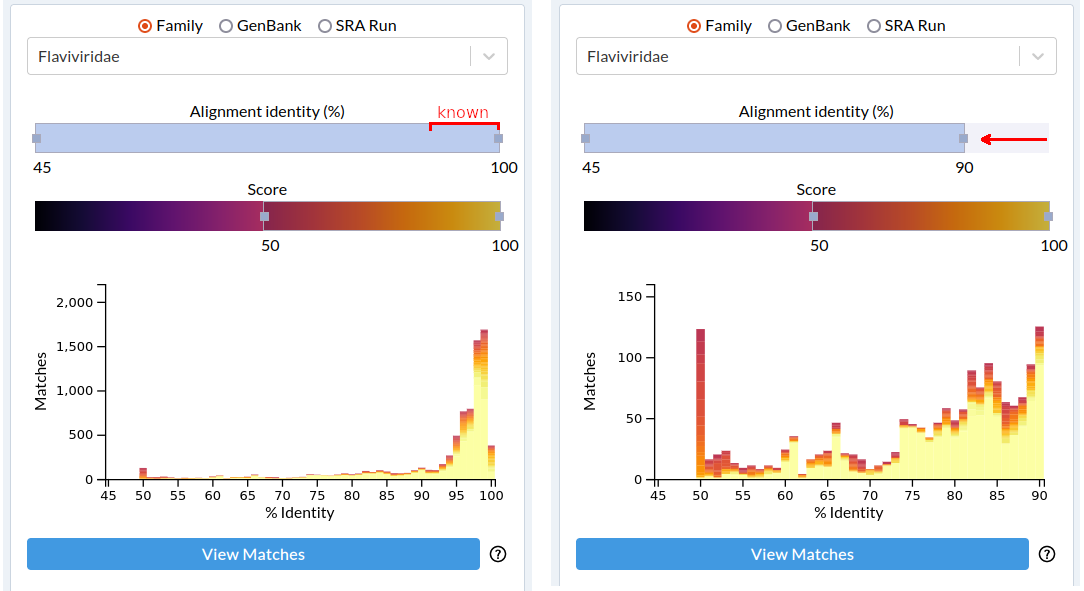
From the drop-down menu, select Flaviviridae. Known viruses will approximately fall in the 90%-100% identity range, so we will focus on the <50%-90% mapped-read identity range with scores of 50-100. Click View Matches.
Note the URL, to reproduce the search results: https://serratus.io/explorer/rdrp?family=Flaviviridae&identity=45-90&score=50-100#result
There are 1,650 sequencing datasets (10 results per page, 165 pages), which match our query. Click Download Matches if you would like a .csv of the results.
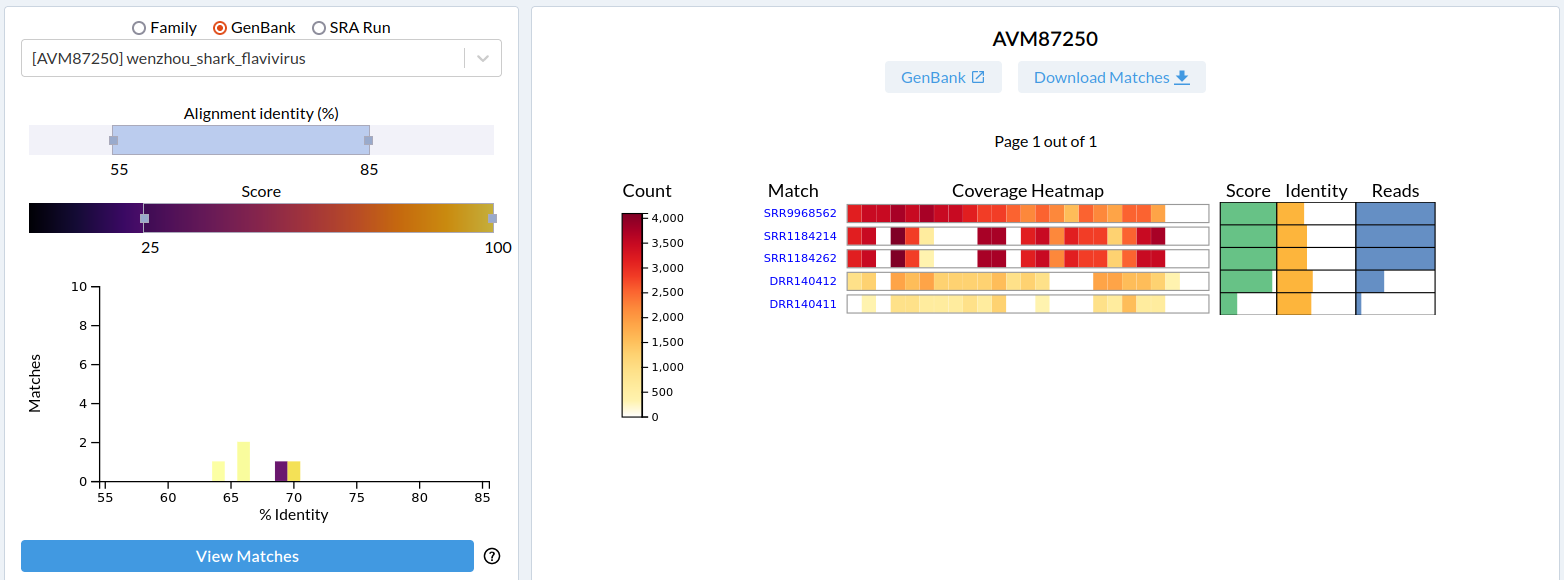
We will select the SRR9968562 dataset, which has a 100 score, 64% read-identity from 17,435 reads. Note that the reads are accumulated in the bins near the sequence C' terminus.
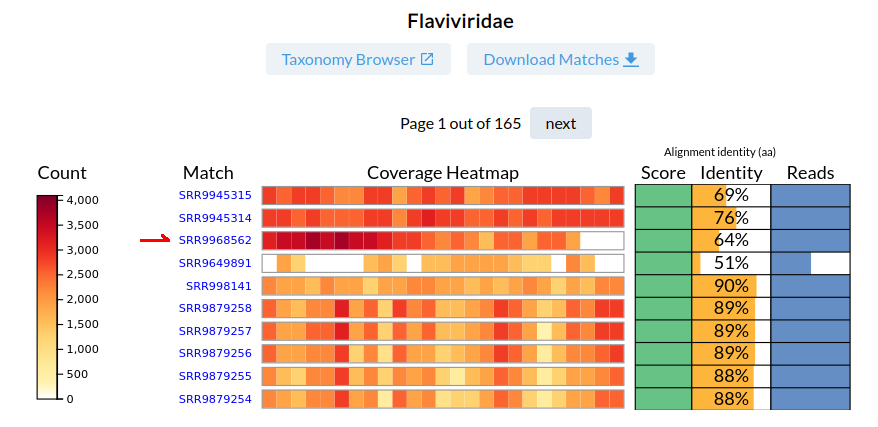
This is a view of all RNA viral reads in SRR9968562. There are matches to two families: 17K reads for Flaviridae, and 84 reads for Permutotetraviridae.
Click on the coverage-heatmap beside Flaviridae to expand the family to the "per sequence" distribution of the reads.
The majority of the reads map to Wenzhou Shark Flavivirus (AVM87250) at 64% identity. A good candidate for a novel virus.
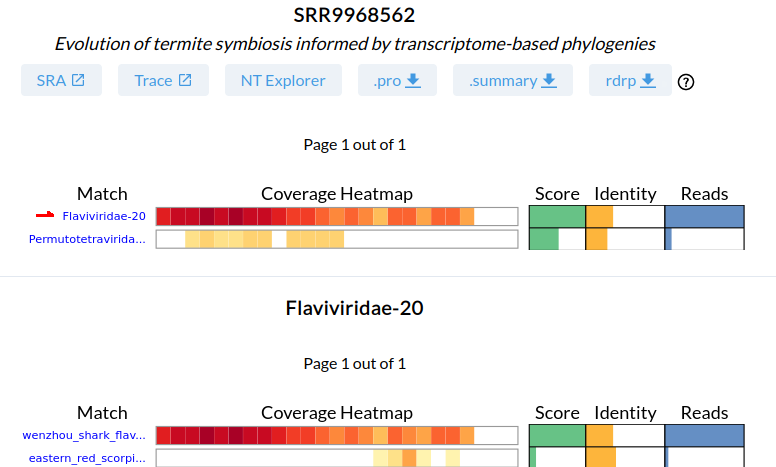
There are 6 links at the top of the page starting with SRA which links to the NCBI-SRA page for this dataset and ends with rdrp which links to the microassembly for this dataset.
All the reads mapped to rdrp1 from SRR9968562 were assembled to yield rdrp-containing contigs, we refer to this as microassembly. Click to download rdrp to download the microassembly. Open this fasta file in your favourite text editor.
The highest coverage contig is (which we know from the summary will be the Flavivirus).
>SRR9968562 NODE_1_length_1481_cov_735.092254
TCTGTGGAATCCAACATCTGCTCTCGTGCCCTCCCAACATACTTTATGTCTTTCATACTG
CTTATGGGTGGCCAGCCTTCCCAAATTTGATTGAATGCTTGAATCATGTTATTGGTGCTC
ATCCAGGGTTGTTCTCTCCACCAGGGGTCCTTGGCTCGACCCATTGGCAATAATCCCCTT
GGTACTATCGCCCTCACCATGTTGGCGGCTAGTCTCACATTCCTCCGTTGTGGGAAAGTG
AGTAATAGGTAGTTGGCATAACCTTTGGCCATGGCTTGTTCAACTTCTCCTGTTTTGTAT
CCACCCAACACTATTCTTGCTTTTCCAAGAATTTCAATCTCACTCCGATCAAGCATTATT
CTGATATCATTTCCTACCTTAACTCTACTGTAGGTGTGGGAACAAAACCATGTGTTCTCG
ATGTTTTTGCTCCATGTTGGTTCTCCAGGTTCAAATTTTAAAACCTTACCGTACTTGGCG
AACGCTGCCACCAGTGTCTCTTCTGGGTGTTGCGTGTCATACAAAACCAGGCAATCGTCC
CCGCTTATCAAGCGGTTCTCTCTTCCATAAGTGTTTCCTAGGTCTTTACTTACGGCTTGC
TGAATGATTGCATTTGTGATGGTGTTCATGGCATAGGTTACTATGGTTCCCGACATTCTC
TGTCCACGACCATGCAACCACATCAGATTTCCGTCTGCGTCCTGCACCCGAATGATGGGG
CTTCTATAGCACTCATAAATGGTCCTAATTTTCCGCCGGTGGTCTTCGCTTTCTGCTCGT
TCCTCACAGATCCGTTGTTCATTTTGCAACTCGTACTCACTCACTCTTGTGTCCCATCCA
GCGATGTCCCCTTGGAACACTTGTCCTTCAGTGGTCTCTCCAGTCATTCTGTTCATCTTT
AGCCGGTTCAAAGCTTTCTCTCCGTACAAGTGTTGGGGTATTCCTGAAACTCCTTCGGGG
GTATTTTCTTCTCCAGCCCAATGGTCTGCATTTAGTGCTCCTAGTGTTCTCATTTCACAT
GCTCGTTCAATCAAATCATAATAAGCTATCATCCTTGAGGTTTTTTGCCCCCACTTATTT
TTCTCCTTTTTCTCTCTTTTTGCCATCGTGTGGAACACTCCTCTCAGGCAAACACCATTT
CTATGGTTTAAGTCTTCAGCTTGAACCTCTCTCCAAAACTCTTCACTTTCCTTCGCTTCC
AGGGCTGTCTTCCATCTGGGATCTCCTACGGCTGCATCATTTCGTATCATCTCCTTCACC
TCGTCCCAACTAAGTTCTCTCAAGGTGGTTGGTAGGGTTTTCTCTACTTCCTTTGAGGCT
TTTTTGAAGTCTTCCATGCCTTCAGGAGGCATATGAGGTGTTGTGTCCACTTTTTCTTTG
AACACTCGCAGCAATCCTACGGGGGAGACGTCTGTTAGCATGTACCTACCAATCCCTGGG
CTTGCTCTCATCAACGGTTCCAGTACCCTATCCCAGATCGG
To test the hypothesis that this is a novel sequence, we will run the BLASTX of this microassembly sequence against the NCBI Non-Redundant Protein (nr) database on their webserver:
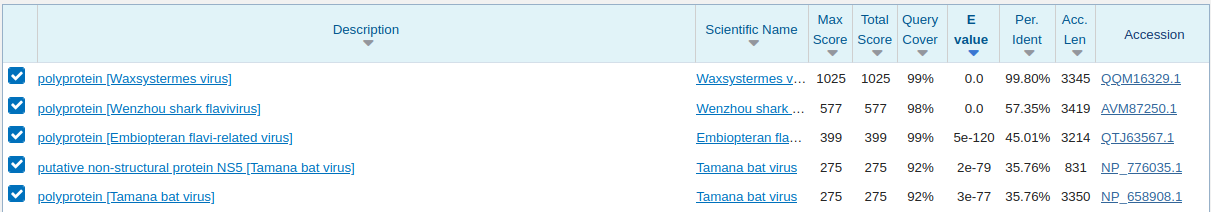
In this case, this sequence had been previously identified by Lay et al., as Waxsystermes virus and deposited on 13-January 2021. The rdrp1 database used in the search was created in December 2020, so this is a perfect positive control, confirming this is in fact a novel virus.
Remember with Serratus we identified 130,000+ novel RNA viruses, and all of these were evalulated against the nr database as being below 90% identity. There's always more viruses to find...
From the last search we identified a "space" containing novel flaviviruses. We can refine our search to find sequences in specifically this vicinity of Flaviviridae.
Navigate back to the RdRP Explorer page and select GenBank as the top-level search.
We know that Waxystermes virus was identified by reads at ~65% identity to Wenzhou Shark Flavivirus (AVM87250). We narrow our search for other libraries with this pattern:
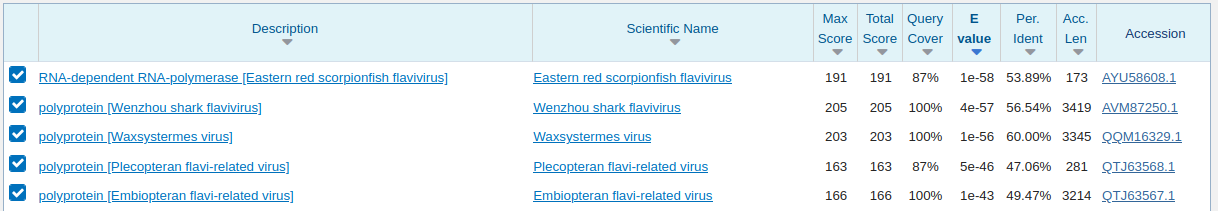
SRR1184214 and SRR1184262 are both from Lepisma sp. (Silverfish), which we can BLASTX. You can see it's 60% to Waxystermes virus, so in the correct vicinity of viruses we were looking for and not reported by Ley et al.
>SRR1184214 NODE_1_length_570_cov_1070.265655
ACGCTGTTCGAGACTTACCGATCACCCATAATAAGACTAAAAACAGAGGGAAATAAAGAG
GTGTTCTTGCATGGTAGAGGACAGCGCATGAGCGGAACAATCATAACGTATTGCAAGAAT
ACTGAGACCAACAGCATCATCCAAGAGTGTGTCTACCGAGAGCTGTTCGGAGAGAGAGGT
GACAGGCGGCGTCGAGCAGTATCAGGAGATGACTGCTGTGTGTGGTGGCCGGAGGAGAGA
ACTCGGGATGCGGTGGAGCAAGCGTTCGCTAAGTTTGGAAAGGTGCTTAAAGACCGGGCT
GGAGAAGGCCACGATTACACCCACAACTTGGAGGATACATGGTTTTGCAGCCACACCTAC
ACTCGTGTCTATGTGGGAAATGGTATCCGGTATCACCTAGATCGTAGCGAAGGAGAGATT
CTGGGAAAAGCTAGACTTGTCCTTGGAGGTTACAAGGATGGAACAGTAGAGTTGGCGATG
GCGAAAGCGTATGCGAACTATCTCTTAGTAACCTTCCCACACAGGAGGAATGTGCGACTT
GCAGCCTTGGCAGTGCGCAGCGTGTGCCCG
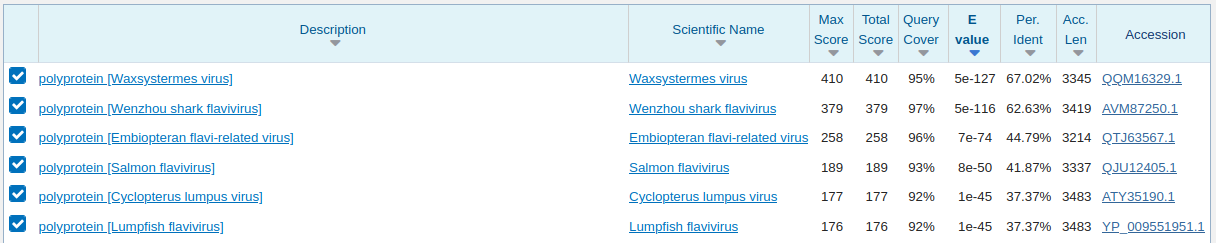
DRR140412 and DRR140411 are from Dianemobius nigrofasciatus, a cricket. Again testing with BLASTX against nr we see this is a novel virus in the correct vicinity.
>DRR140412 NODE_2_length_892_cov_10.260973
CCGATCTGCCTTTTTTCTTGCAGTTTTCTTATCACACACTGTTGGATCACTGCATTGGTG
ATAGTATTCATGGCATAAGTAGTCACGCTCCCCGACATTCTCTGGCCCCTGCCATGCAGG
TACAAGTCGTATCCTTGTCCACTTCTGAGCTTTACGATGGGGGATCGGTAACACTCATAA
ATCGTCCTAATGGCTCTCCGATGGCCTTTGCTCGACGCTCTTTCTTCGCAGATTCTCTGC
TCATTCTCTAGCTCAAACTCACTCACACGCGTGTCCCAACCACCTATGTCGCCCTGGAAC
ACTACTCCTCCCGGGATTTCCCCCGAGTTCCTGTTGGCCCCCACCCGGCTAGCTATCTCT
TCCCCATACATGTGTTGCGGAATACCCGTAACACCTTCTGGCAAGTTTTCCTTAGCTGCC
CAGTGGTCTACGTTTATAGCTCCTAGGGCCTTCATTTCACAAGCCCTTTCCACCAAGTCG
TAGAATGCTATCATCCTGCTAGTTTTTGGCCCCCATTTATTCTTCTCCTTCTTCTCTCTC
TTTGCCATAGTGTTGAAAATCCCTCTCTTGCAAATTCCATTCACATGATTTTCGTGCTCT
AAGGCGCATTCATTCCAAAAAGCGGGGTCTTCTTTGGCTTCCTTGGCTGTCGTCCACCGG
GAGTCTCCCACTGAGGCGTCATTCCTGATACAGTCTTTCACCTCGCTCCAGGTCAACTCC
CTTAGGATTTTTGGCAATCCACTCTCAATTTCCCTTGAACACTCCCTGAAAAGGGCCCTG
CTCTCTTTGGGCATGTATGGAGTTGTGTCGATTTTTTCCTTGAAGACTCTCATCAAGCCT
TCATCACTCACGTCTGTTAGTGTATAACGCATGATCTTCTGGTTTACTTTAG
That's it! You've used the Serratus platform to uncover novel viruses.
Here we have provided an overview on how to apply the serratus.io web interface to easily "click and discover" novel viruses. As you interface the data more, you will grow an intuition for paths to identifying novel viruses and what they "look like".
In all forms of analysis, this is by far the fastest and easiest way to reach the relevant data. Even when doing systematic analyses as outlined in further tutorials, we come back to the steps outlined above as a sanity check and means of confirming the validity of what a larger analysis is telling us.
- Novel Bornaviruses in colubrid and viperid snakes is great example of the next steps to turn your findings into a short paper
- Serratus.io basic overview
- Accessing the
SQLdatabase directly -
rdrp1reference query sequences - Understanding sources of error for alignment-based analysis
- Tutorial: Running DIAMOND nr search
- Tutorial B: RdRP palmprint sequence-search
- Tutorial C: RdRP microassembly sequence-search