This forked repository uses a modified PP-Seq algorithm (point process model of neural sequences) dubbed "PP-Almost-Seq" to detect repeated firings of polychronous neuronal groups (PNGs) in data and detect the next-best-candidate sequences. PNGs are collections of neurons that fire with a specific order and delays and are theorized to store memories in the brain. Currently, the code focuses on stress-testing the algorithm on multiple instances of planted sequences. Research endeavors are documented here. To learn more about my research, you can also peruse the slides at the top of the documentation or check out this poster:
All major PNG-related files are in the folder labeled izhikevich_pngs. One can use the theDataMachine Python file to construct a network of neurons using the simulator and export the spike data into a suitable format for PP-(Almost-)Seq. The png and autoPng files use the PP-Almost-Seq algorithm to cluster spike data into sequences. autoPng allows the user to analyze multiple spike data files in one runthrough, and both save the resulting graph as a png (the image file format) in the graphs folder. For information on how to use autoPng and png, check out the songbird demo folder provided by the original authors.
The prototype PP-Almost-Seq code can be found in the gibbs.jl file. The author plans to clean up and sort the currently extremely long dictionary output. In the theDataMachine file, there are multiple functions to trigger sequences in data with various instances and delays. There is also a section of code that allows one to embed PNGs in the network via network connectivity and weights.
From the original readme:
This repo implements the point process model of neural sequences (PP-Seq) described in:
Alex H. Williams ☕, Anthony Degleris 🌄, Yixin Wang, Scott W. Linderman 📢.
Point process models for sequence detection in high-dimensional neural spike trains.
Neural Information Processing Systems 2020, Vancouver, CA.
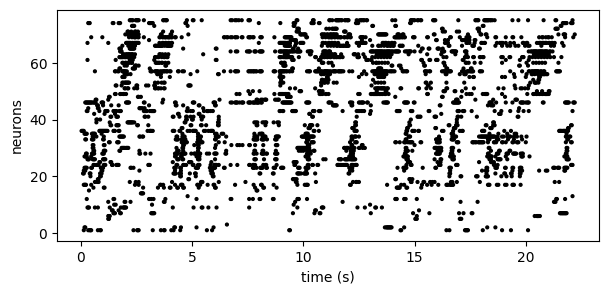
This model aims to identify sequential firing patterns in neural spike trains in an unsupervised manner. For example, consider the spike train below(1):
By eye, we see no obvious structure in these data. However, by re-ordering the neurons according to PP-Seq's inferred sequences, we obtain:
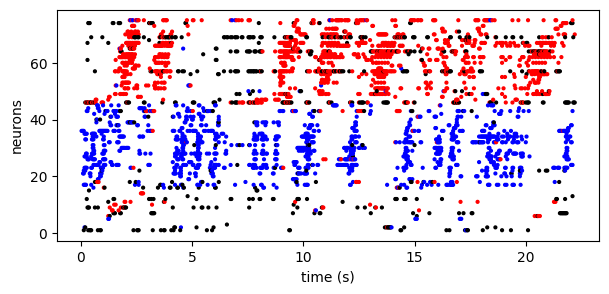
Further, the model provides (probabilistic) assignment labels to each spike. In this case, we fit a model with two types of sequences. Below we use the model to color each spike as red (sequence 1), blue (sequence 2), or black (non-sequence background spike):
The model is fully probabilistic and Bayesian, so there are many other nice summary statistics and plots that we can make. See our paper for full details.
Footnote. (1) These data are deconvolved spikes from a calcium imaging recording from zebra finch HVC. These data were published in Mackevicius*, Bahle*, et al. (2019) and are freely available online at https://github.com/FeeLab/seqNMF.