forked from r4ds/bookclub-rpkgs
-
Notifications
You must be signed in to change notification settings - Fork 0
/
Copy path05-fundamental-development-workflows.Rmd
204 lines (125 loc) · 9.24 KB
/
05-fundamental-development-workflows.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
# Fundamental development workflows
**Learning objectives:**
- When and how to make an R package.
- RStudio projects.
- Why not to mess with your working directory.
- Test driving code.
## Survey the existing landscape
- Be sure that the package doesn't overlap *too* much with existing packages:
* Is there an existing package that does what you need?
* Could you contribute to a similar, existing package to add the functionality you need?
[List of packages on CRAN](https://cran.r-project.org/web/packages/available_packages_by_name.html)
- Payoffs:
* Product: your life will be better when this functionality is implemented formally, in a package.
* Process: greater mastery of R will make you more effective in your work.
## Considerations
Ensure that the package fits in well within the domain of existing R packages.
For modelling packages, using the [hardhat](https://hardhat.tidymodels.org/) package helps the developer use best practices and ensures that the resulting package can be used within the tidymodels ecosystem (and get used more).
You may also want to include consider including appropriate methods to be consistent with other related, packages. For example, many modelling packages have predict, plot, and summary methods, and users may expect this functionality from a new package that you create.
## Naming your package
See the naming convention used for existing packages:
[List of packages on CRAN](https://cran.r-project.org/web/packages/available_packages_by_name.html)
Laws:
1. The name can only consist of letters, numbers, and periods, i.e., ..
2. It must start with a letter.
3. It cannot end with a period.
(no underscores `_` or hyphens `-`)
Recommendations:
1. No periods.
2. Easy to Google.
3. Avoid confusion with an existing R package (not on CRAN/Bioconductor/GitHub) or well known package from other language (numpy, pandas)
4. All lower case if possible.
5. Abbreviations to reduce name length (brms = Bayesian Regression Models using Stan)
6. Adding in capital R's for fun.
7. Not getting sued (~~facebook~~ Rfacebook, ~~Dropbox~~ RDrop, ~~apple~~ appler)
[Here](https://www.njtierney.com/post/2018/06/20/naming-things/) is a fun blog post on naming R packages by Nick Tierney. Also recommend checking out his package `greta` which is really cool and has useful example of using python (to access tensorflow) from R in a package. He also has a guide on [renaming an R package](https://www.njtierney.com/post/2017/10/27/change-pkg-name/)
## Creating a new package
- Call `usethis::create_package('yourPackageName')`
- In RStudio, do File > New Project > New Directory > R Package. This ultimately calls usethis::create_package(), so really there’s just one way.
~~(I prefer to use RStudio - show using create_package from bash)~~
### What does this do?
This creates your project structure for you:
- `R/` directory
- `DESCRIPTION` file
- `NAMESPACE` file
- `pkgName.Rproj` file
- `.Rbuildignore` & `.gitignore`
DO NOT use `package.skeleton()` - it creates a package which will lead to errors when running `R CMD build` and is not consistent with the devtools style of development.
### Where to put this package?
This is distinct from where your R packages are installed. It makes sense to put this in a similar area to where you keep your other R projects.
Within my `/user/documents/`, I have `R_projects/` and `R_pkgs/`
## Using RStudio Projects
You can use devtools without using RStudio and visa versa, but it's not as nice.
### Benefits of RStudio Projects
- Each project is isolated and code is contained within the project.
- Easy to start up a project in a fresh instance of RStudio.
- Helps mitigate workspace overlap (e.g., function masking and overwriting accidentally same-named objects)
- Keyboard shortcuts for common code development tasks (build, load, test, document, check)
* alt-shift-k for shortcuts
### Make an R Project for an existing package
If you already have an R package which isn't within an R Project, you can:
- In RStudio, do File > New Project > Existing Directory.
- Call `usethis::create_package()` with the path to the pre-existing R source package.
- Call `usethis::use_rstudio()`, with the active usethis project set to an existing R package. In practice, this probably means you just need to make sure working directory is inside the pre-existing package.
(using `usethis::use_rstudio()` from bash fixes the problem of the other not being created)
### R Project tidbits
A directory for an R Project will have a `.Rproj` file, typically with the same name as the directory (and the package if the project is a package). It's best case for all of these names to be the same.
This file is just a text file and you don't need to modify it by hand. It's possible to modify it with 'Project Options...' in RStudio (drop down on top right).
Launch an R Project by double clicking the `.Rproj` file or from within RStudio.
## RStudio, `usethis` and working directories
`usethis` functions don't take a path (i.e. `usethis::use_r("new_fx")`)
`usethis` assumes that the following coincide:
- The current RStudio Project.
- The active usethis project.
- Current working directory for the R process.
It's therefore much easier to not mess with your working directory for your R console. Leave it as the project directory so that when you're calling functions from `usethis`, it puts things in the right place.
You can double check that these three directories are coinciding by running:
```{r}
usethis::proj_sitrep()
```
Similarly, when you run helper functions from devtools to develop your package (`document()`, `check()`, `test()`), these are also expecting your working directory to be the project directory. The also expect that your project follows the conventions of project structure (test scripts living in `tests/`, R code living in `R/`, vignettes living in `vignettes/` compiled code living in `src/`, images living in `man/figures/` etc).
## Test driving a package
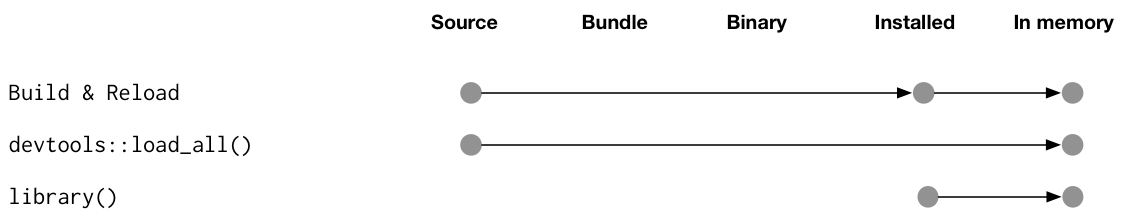
### Using `load_all()`
Running `load_all()` is the fastest way to re-load the functions in your package -- much faster than installing it (`install.packages(pkgName)`) and loading it into memory `library(pkgName)`. **This is helpful for iteratively updating or tweaking functions you're working on**.
### Using `test()`
Running `test()` runs `load_all()` and then the test files that live in `tests/testthat/`. If you have a test file which captures the behaviour in the function that you desire, you can quickly assess whether the changes you make in the function have worked by running `test()`.
### Coping with being impatient
`check()` can take a long time and leave your console occupied. A solution to this is to run using the build tab (ctrl-shift-E) or as a separate job:
`job::job({check()})`
To test examples in your documentation without running `check()`:
`run_examples()`
### (Previous speaker's) bad habit
I like to have a `working_space.Rmd` in the package directory which I use to test package code away from the console. You'll get a NOTE on the check:
> N checking top-level files ...
>
> Non-standard file/directory found at top level:
>
> 'working_space.Rmd'
But you could also add this to `.Rbuildignore`/`.gitignore`.
It can be tricky to incorporate everything into a test script, and sometimes you may want to have a closer look at what's going on within a function. This can more easily be done by throwing some earlier `return()`s into your package functions, run `load_all()` to make them effective and then running code in your `working_space.Rmd` file.
## Meeting Videos
### Cohort 1
`r knitr::include_url("https://www.youtube.com/embed/DcV2km3Qx-g")`
### Cohort 2
`r knitr::include_url("https://www.youtube.com/embed/gr3vhN4XxWI")`
### Cohort 3
`r knitr::include_url("https://www.youtube.com/embed/Hg37VT-cWVM")`
<details>
<summary> Meeting chat log </summary>
```
00:25:44 Ryan Metcalf: Mac: Opt->Shift->k
00:32:09 Ryan Metcalf: The first link wasn’t credible. There is a `{changer}` additional add in, that may aid in naming convention change. https://cran.r-project.org/web/packages/changer/changer.pdf
00:34:36 Isabella Velásquez: here’s an article (by Nick Tierney!) on how to change package names (note, it is from 2017): https://www.njtierney.com/post/2017/10/27/change-pkg-name/
00:57:23 Isabella Velásquez: https://twitter.com/jayyqi/status/1228049523806617601?ref_src=twsrc%5Etfw%7Ctwcamp%5Etweetembed%7Ctwterm%5E1228049523806617601%7Ctwgr%5E%7Ctwcon%5Es1_&ref_url=https%3A%2F%2Fivelasq.rbind.io%2Fblog%2Fyou-cran-do-it%2F
01:01:12 Brendan Lam: This is helpful^ Thanks!
01:01:44 Isabella Velásquez: Is this what you are referring to? RMarkdown Driven Development (RmdDD) https://emilyriederer.netlify.app/post/rmarkdown-driven-development/
01:03:25 collinberke: Yes!
01:04:04 collinberke: https://www.youtube.com/watch?v=eMWgu9OQ0m8&list=PL3x6DOfs2NGiXMln8bxY7e0XgA5z1cp-8&index=4
01:04:24 Isabella Velásquez: Thanks!
01:06:39 Ryan Metcalf: https://www.rstudio.com/conference/
01:06:49 Isabella Velásquez: HAVE to
01:10:21 Isabella Velásquez: Here’s a video of Emily’s talk from a conf in the past : https://www.youtube.com/watch?v=B6YO5nW46SU
01:16:22 Ryan Metcalf: `git checkout -b <name of your branch>`
```
</details>